Morphological analysis of tissues
Quantitative histology to measure the development of organs
The use of the image analysis in developmental biology allows to monitor morphological parameters through the development of organs and tissues. The following examples illustrate for mice the development of mammary glands in the embryonic phase and to the adult.
Structure of mammary buds
Formation of buds is measured according to the following diagram.
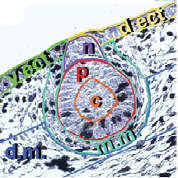
- Dorsal ectoderm (d.ect., vert)
- Ventral ectoderm(v.ect., jaune)
- MR core (c, orange)
- MR periphery (p, rouge)
- MR neck (n, violet)
- Mammarymesenchyme (m.m., turquoise)
- Dermalmesenchyme(d.m., bleu)
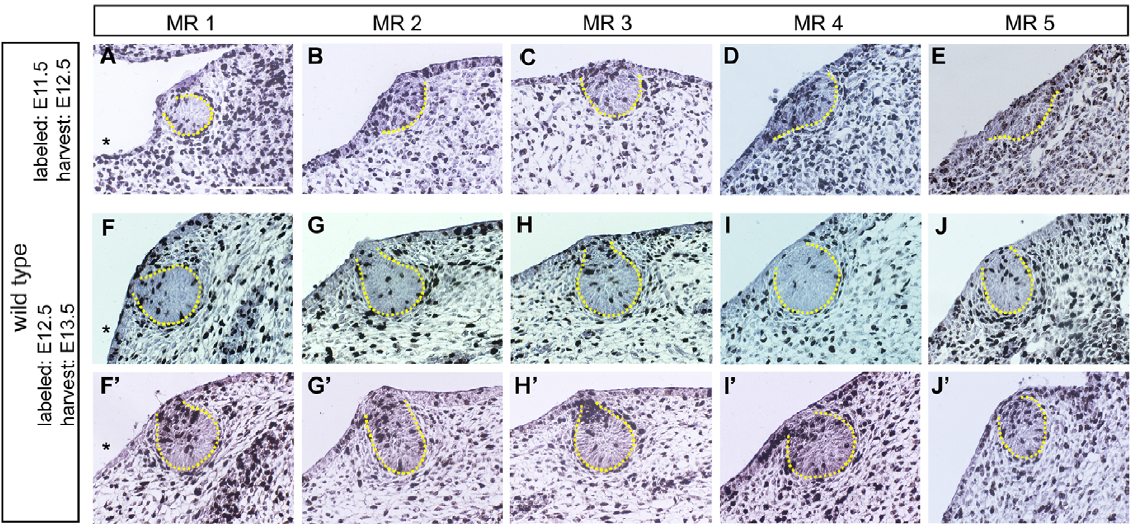
Effects of genetic mutations in mammary buds
5 pairs of mammary buds (MR1 to MR5) between the 12th and 14th embryonic day are analyzed at mutant mice Gli3Xt-J/Xt-J and compared with wild mice.
Implementation of a specific software for automatic image analysis
A protocol of image analysis of based on Metamorph journals ( Molecular Device Corp.) and on Matlab scripts (MathWorks) allow:
- De définir d’une manière semi-automatique
- bud area (p+c)
- neck (n)
- Automatically define all areas (d.m. m.m. c, p, v.ect, d.ect)
- Visualize segmentation steps for QC (quality control)
Tools of image processing involved in this study are :
- Mathematical morphometry
- Analysis based on wavelet transforms
- 3D reconstruction
- Active contours
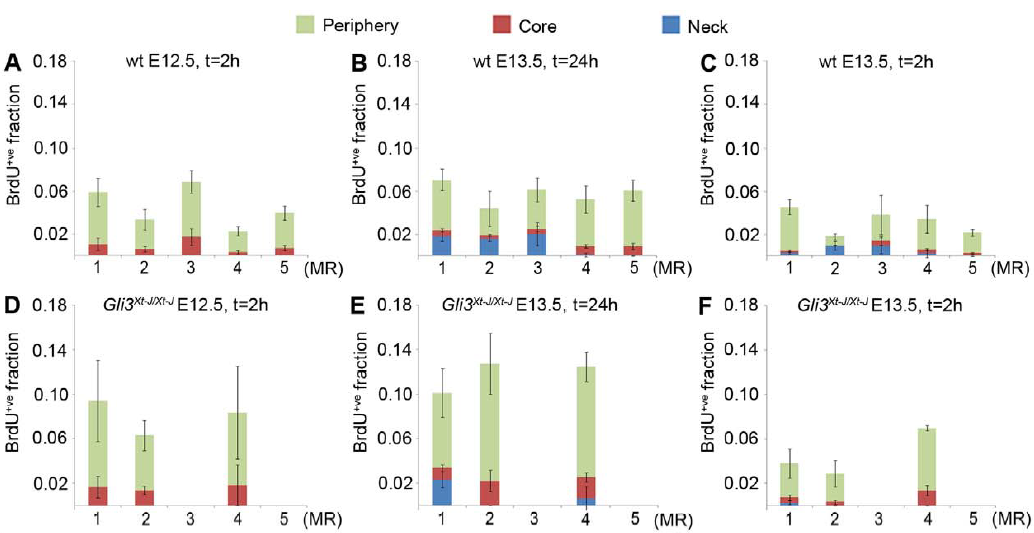
Statistical analysis of the morphological parameters during the development
The following figure shows the evolution of the rate of positive cells BrdU+ve (proliferation marker) in the sub-regions of the bud (core, periphery and neck)
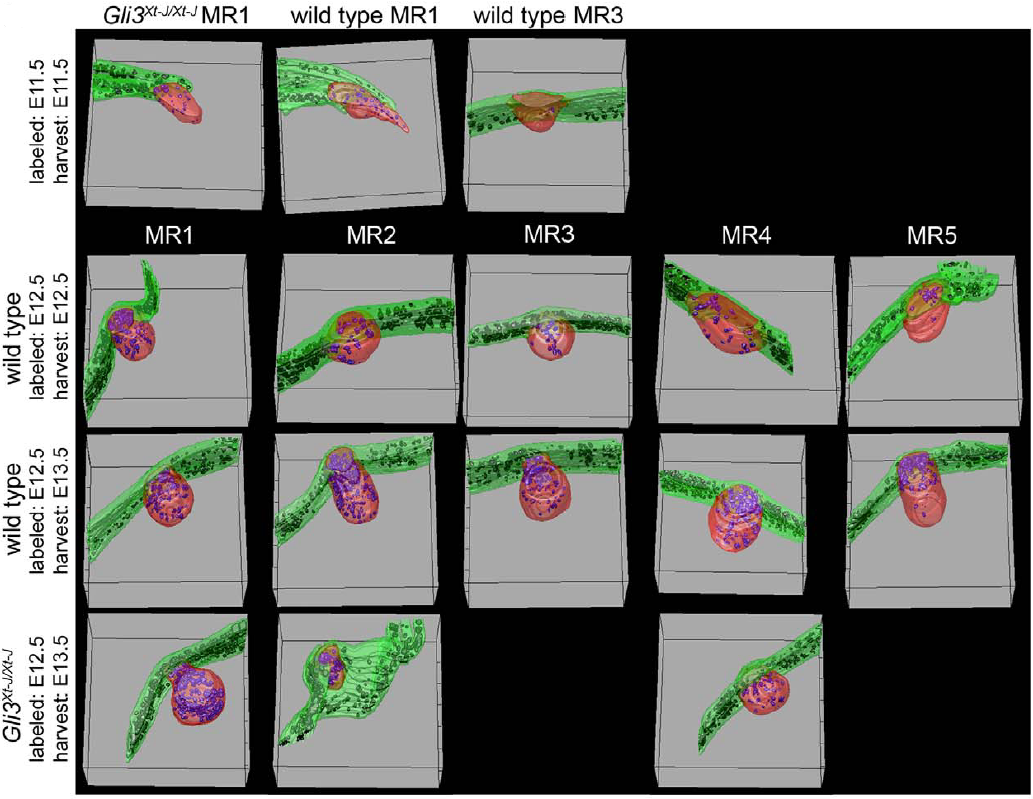
3D quantitative analysis in histology
Analyses were driven on successive slices allowing 3D visualization of buds (in red), positive cells BrdU+ve (in black) and skin (in green)
References
Ectodermal influx and cell hypertrophy drive early growth of all five murine mammary rudiment pairs, and are differentially regulated among them by Gli3 (2011). May Yin Lee, Victor Racine, Peter Jagadpramana, Li Sun, Weimiao Yu, Tiehua Du, Bradley Spencer-Dene, Nicole Rubin, Lendy T. Le, Delphine Ndiaye, Saverio Bellusci, Klaus Kratochwil, and Jacqueline M. Veltmaat.
PLoS ONE 6(10):e26242 1-15. PMID:22046263, Epub: 2011 Oct 27; pdf.
Interested in softwares or services?
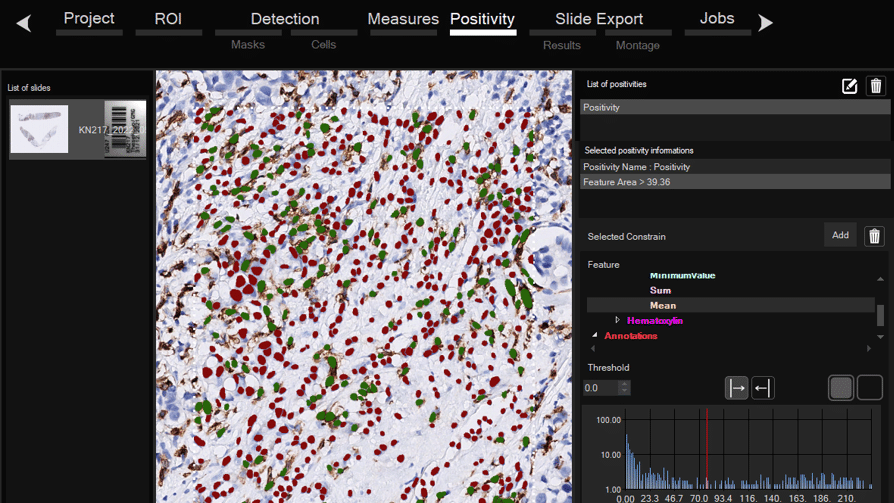
Software for histological samples and TMA
Automatically detects and quantifies your histological slides easily, without requiring bioinformatics expertise. IHC/Fluorescence/Deep learning
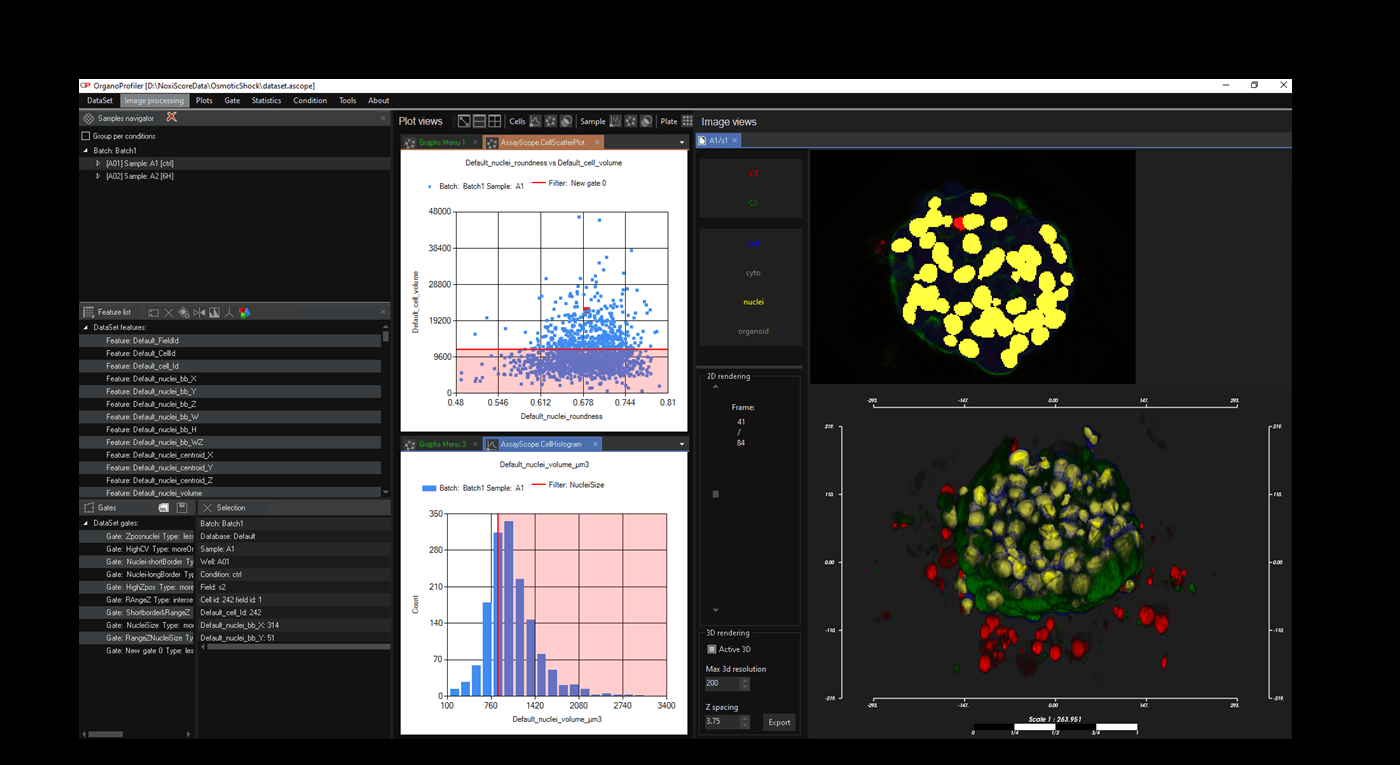
Software for organoid, spheroid and 3D cell culture and tissue
Automatic 3D segmentation and high-content screening analysis software to navigate your assays and monitor drug effects
Services of analysis and characterization
- Image analysis in patients studies
- Characterization of drug/compounds effects
- Automatic detection of rare events

Software development for biomedical imaging
- Customized software solutions with ergonomics UIX
- AI for automatic analysis
- Industrialization of analyses in laboratories
QuantaCell, Hôpital Saint Eloi, IRMB
Contact
+33 (0) 9 83 33 81 90
80 av Augustin Fliche
34090 Montpellier, France